Response format
The response format is tailored to each specific endpoint. To view the response format for a particular endpoint, please visit our Swagger UI. Once there, select the endpoint of interest and scroll down to the “Responses” section. A ‘200’ response code indicates a successful request, while any other code signifies an error.
Endpoints typically support returning data in JSON, CSV or TSV, with JSON being the default.
Genomic sequences (such as those from unalignedNucleotideSequences, alignedAminoAcidSequences, etc.) are provided in the
FASTA format by default,
but can also be requested in JSON or NDJSON format.
Example
Section titled “Example”To understand the response of the nucleotideMutation endpoint, refer to the relevant section in the Swagger UI:
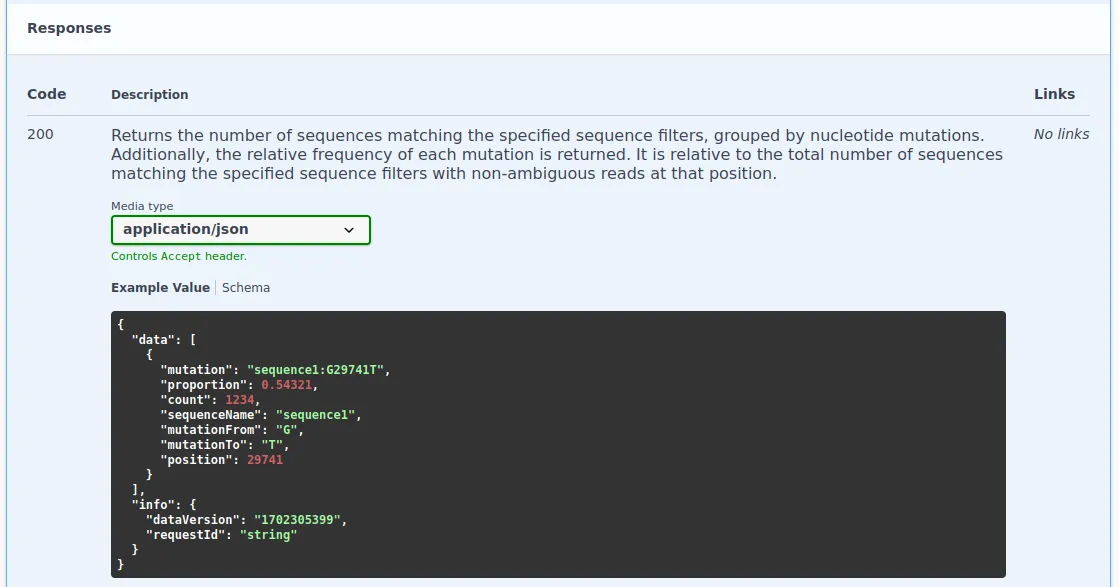
At the top, you’ll find a plain text description of the response. The middle section displays a JSON format response example. For detailed information about the response data, click on ‘Schema’. This reveals the response structure and descriptions for each field, accessible by clicking the three dots next to them.
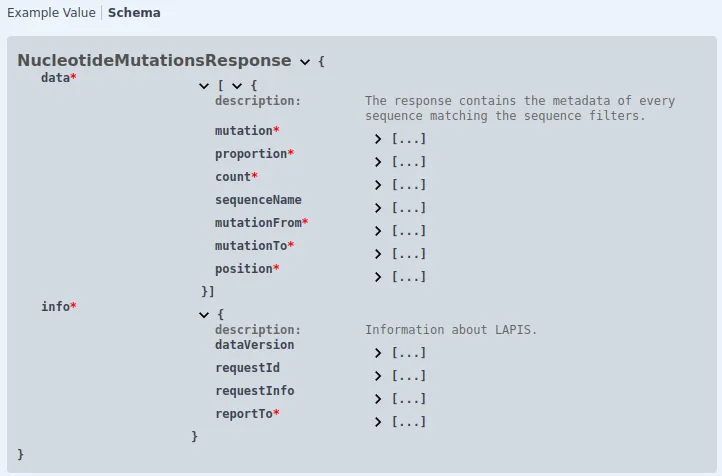
Fields marked with an asterisk (*) are always included in the response; unmarked fields may be omitted.